5,500 new RNA virus species discovered in the Ocean using samples collected during Tara Oceans mission
Using samples collected during Tara’s voyage, a team of researchers have discovered thousands of new species of RNA viruses in the ocean. Analysis of RNA viruses from across the global ocean marks a new chapter in understanding RNA virus diversity, evolution, and ecology.
Discovering the RNA virosphere
The ocean was once thought to be a vast desert in terms of viruses. Today, scientists are turning to the multitudes we now know inhabit the world’s seas to unravel mysteries of their biology, evolution, and function.
While viruses are synonymous with disease, they also infect smaller organisms that run the Earth, like bacteria and plankton. They are a natural and vital part of the Earth System.
By applying new technologies and approaches to samples collected during the Tara Oceans expedition (2009-13), our knowledge of viruses, and their surprising roles in ecosystems, evolution, and life is rapidly advancing.
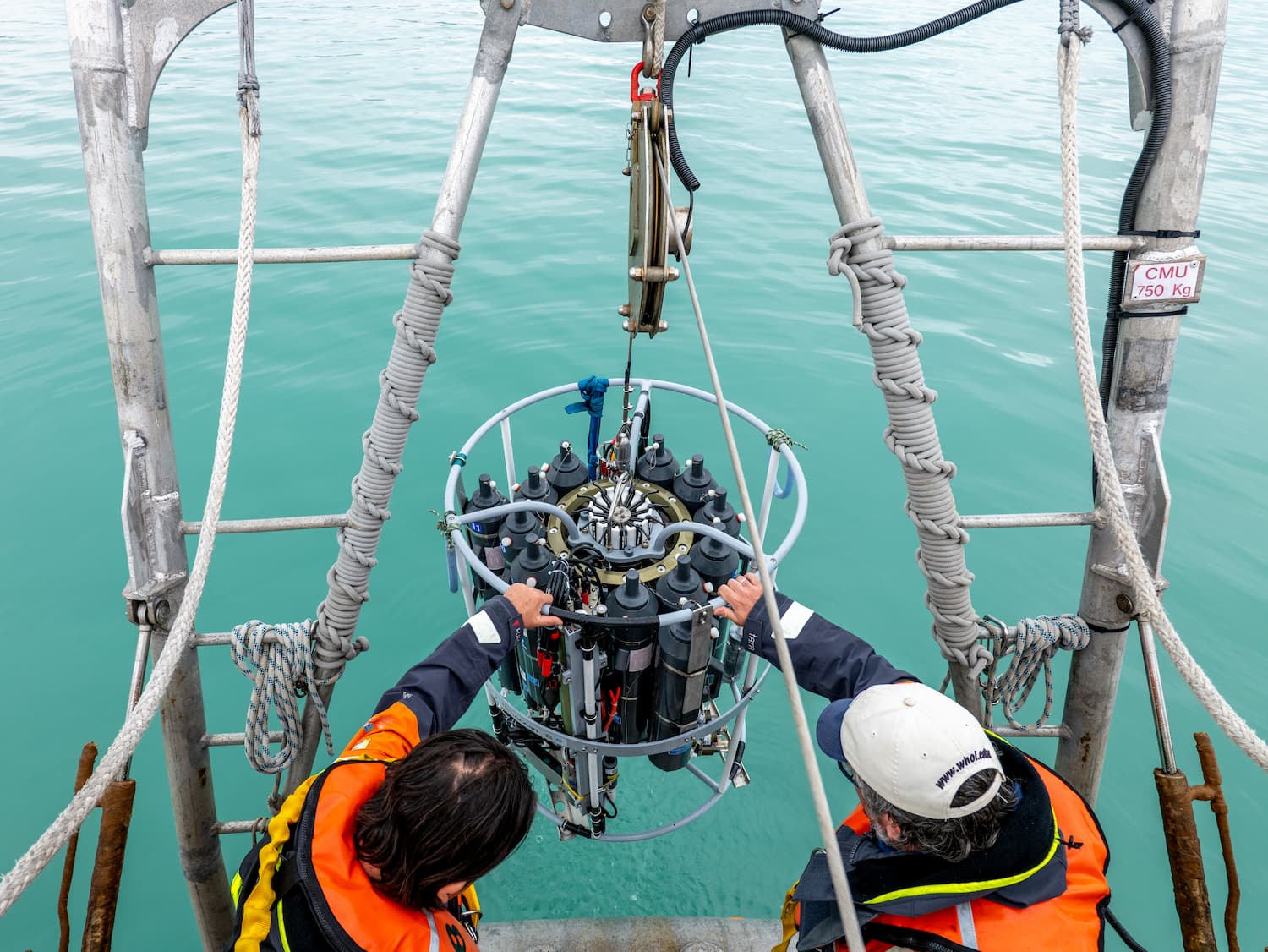
DNA and RNA marines viruses
However, until now, most of the ocean-dwelling viruses categorised were DNA viruses – especially bacteriophages, viruses that infect bacteria. But it has long been known that RNA viruses – typically infecting eukaryotes such as plants, fungi, protozoans, and animals – are also abundant in the global ocean.
This left open the distinct possibility that many more viral discoveries lie in wait.
New scientific discovery on RNA viruses
Now, using samples collected during Tara’s voyage, a team of researchers have discovered thousands of new species of RNA viruses in the ocean. Their findings, published in Science on 8 April, transform our understanding of RNA viruses and their roles in nature.

In particular, the new study:
- Revises the RNA virus ‘tree of life’ ;
- Doubles the number of known phyla of RNA viruses – including two new phyla that can be found across the global ocean ;
- Reveals 5,000 new RNA virus species ;
- Shines light on the role of RNA viruses in the evolution of early life on Earth ;
- Develops new machine learning approaches that can support the identification and classification of RNA virus species ;
- Points to a key role of RNA viruses in essential ecosystem services such as oxygen production and ocean carbon capture.
A study initiated by the Tara Ocean Foundation
The new study was initiated by the Tara Ocean Foundation and the Tara Oceans scientific consortium. The work utilised improvements in sequencing technologies led by the French National Sequencing Centre (Genoscope) and the French Alternative Energies and Atomic Energy Commission (CEA).
Combined with viral reference catalogues, these advancements provided a platform to take snapshots of RNA virus populations across the Atlantic, Pacific, Arctic, Southern, and Indian oceans.
The work furthers understanding the nature and diversity of RNA viruses, where they live in the global ocean, their surprising roles in carbon capture and oxygen production, and their influence on the evolution of all life.
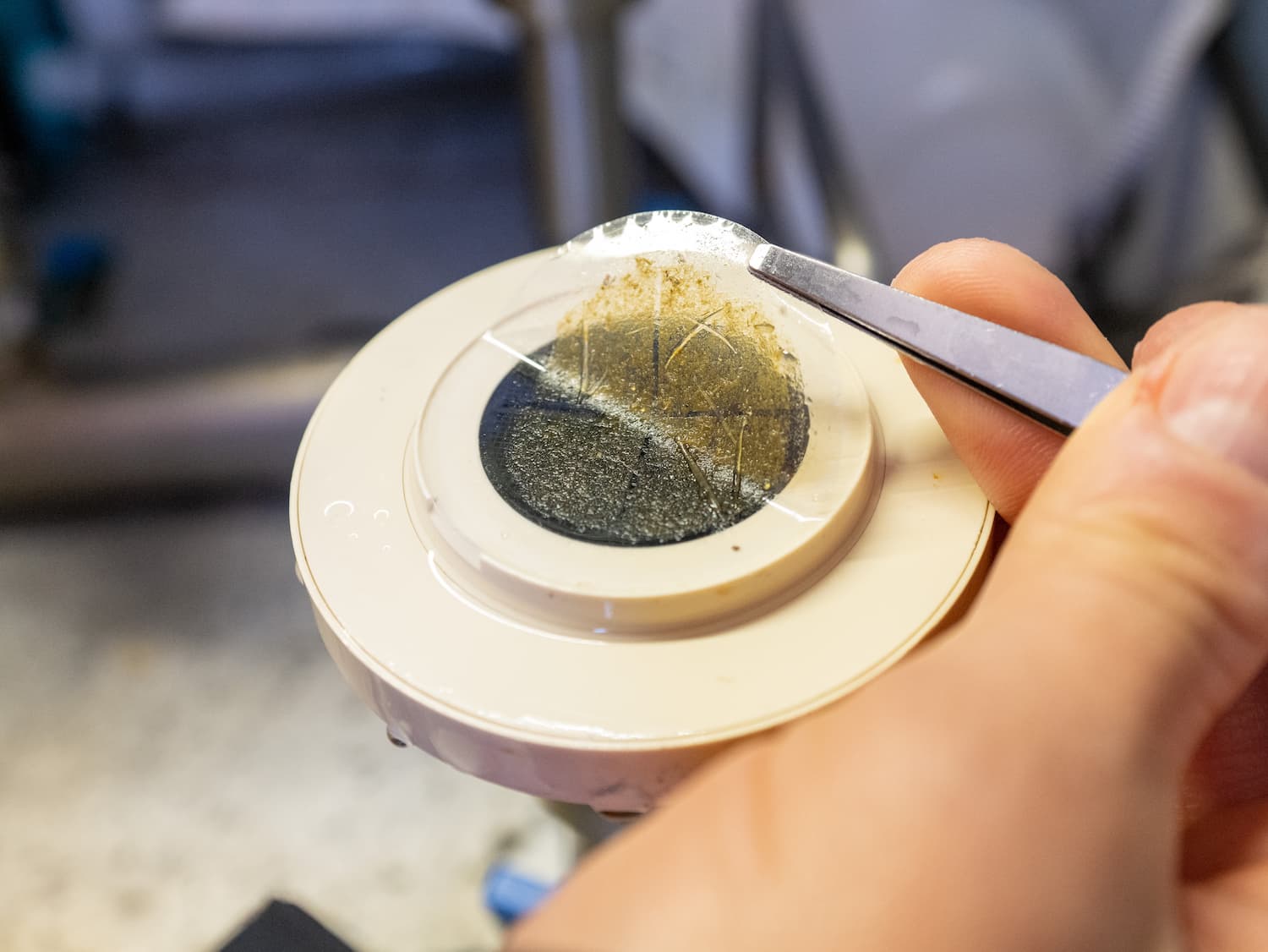
UA new window on RNA virus diversity and evolution
When Charles Darwin sketched an early tree of life in his notebook, it helped to visualise the ancestral relationships between species. Ever since, scientists have used trees to map out the relationships between biological entities to help develop a framework for understanding nature.
In the animal world, major groups of animals, or phyla, include arthropods – such as insects and crustaceans; annelids – for instance leeches and worms; and chordates –like humans and fish. As researchers learn more about viruses in the global ocean, they want to map them in similar ways.
But unlike cellular species, viruses have no universal set of genes. Therefore, when pieces of genetic material are isolated from an unknown virus, it is hard or even impossible to work out who or where it has come from. Scientists have aimed to make sense of this uncertainty using methods that enable them to bring together genome segments, identify overlapping pieces, and piece together viral DNA like a jigsaw.
Techniques such as metagenomics have recently transformed our understanding of DNA viruses. However, a major gap in our understanding was that of RNA viruses, whose genetic information is not typically revealed in metagenomic studies. This meant that no one had really looked at RNA viruses in the global ocean in a systematic way before.
Metatranscriptomics, a technique adapted to the analysis of RNA molecules
The researchers turned to metatranscriptomics – a burgeoning method that enables the study molecules of RNA in the ocean (or any other) environment. Metatranscriptomics has been used in the rapid identification of pathogens underlying diseases such as Covid-19. And now researchers have applied it to water samples collected across the global ocean during Tara’s five-year voyage.
Amongst a horde of headline findings, the study has doubled the amount of known RNA virus phyla, from five to 10. Amongst these highly diverse viral phyla, the research team have identified 5500 new species of RNA viruses. The findings revise the RNA virus tree of life and hold the potential to revolutionise our understanding of their biology.
The analysis unmasks roles played by RNA viruses in the evolution hundreds of millions or billions of years ago. Combining a range of computational approaches, including deep phylogenies, the new study revealed that some of the most abundant and widespread RNA viral groups identified were played a key role in the evolution of early life on Earth.
Water samples taken during the Tara Oceans expedition were taken systematically across the global ocean: the research also shines light on where particular virus phyla are likely to be found in the world’s seas. Two of the new phyla identified by researchers are abundant across the global ocean.
The study also developed new machine learning approaches to make sense of the soup of viral RNA found in the waters. The new method pre-filters highly divergent RNA sequences into clusters so that they can be studied in a more objective and semi-automated way than was possible before. This helps to overcome technical barriers for identifying, describing, and classifying virus species. Such steps are critical for cleaning up controversy and challenges in RNA virus discovery work.
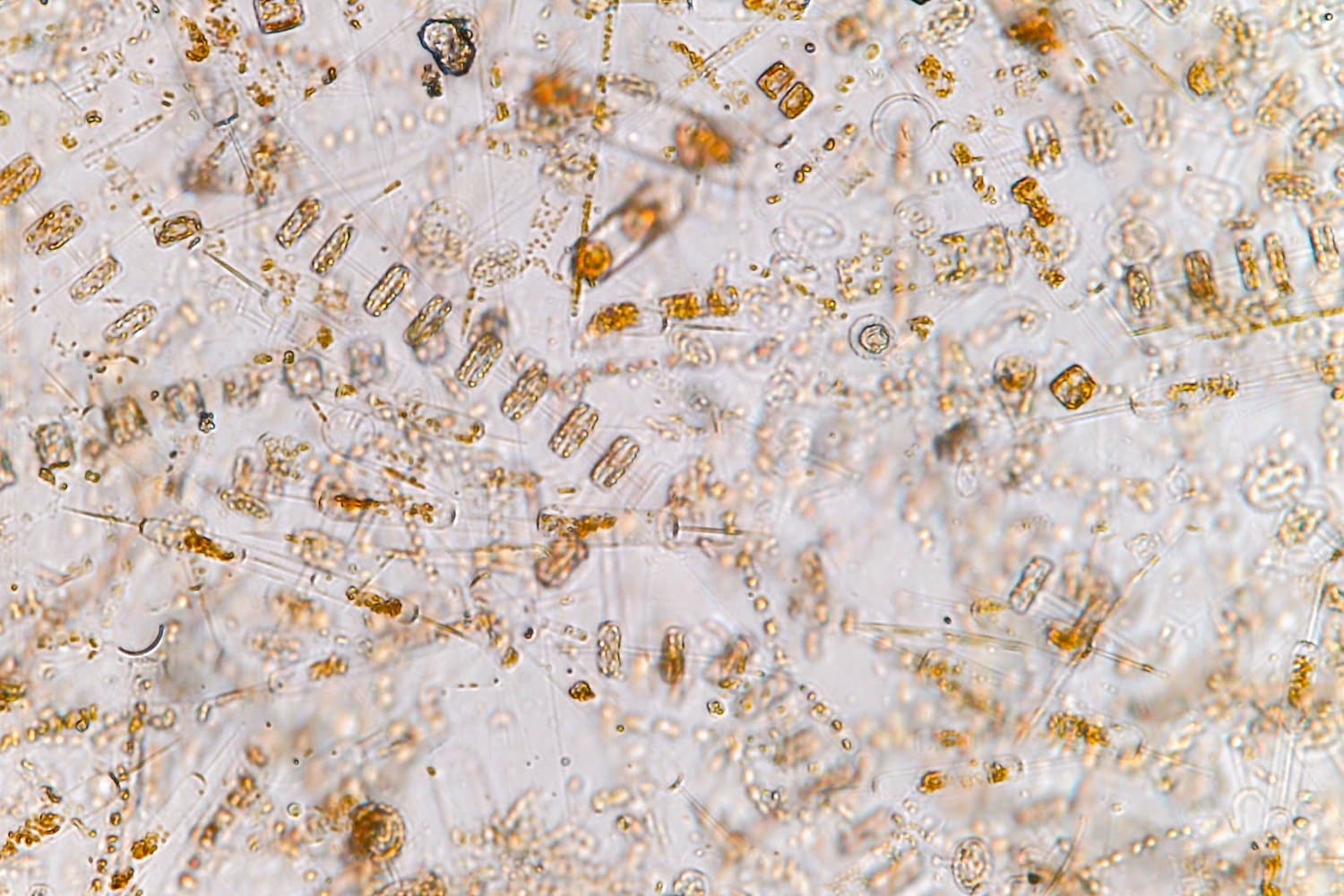
RNA viruses play a major role in ocean ecology
Plankton make up 70-90% of the total biomass in the ocean and capture the energy that sustains all ocean life – from crabs, to barnacles, to fishes, to octopuses, to whales. Their communities create an immense system of organic matter that forms the base of the ocean food web, bringing essential nutrients such as carbon, potassium and iron into the global food chain.
During primary production, microalgae and photosynthetic bacteria absorb carbon dioxide, change it into living matter, and collectively produce at least half of the total oxygen in the atmosphere. Plankton are also responsible for roughly half of all global photosynthetic carbon dioxide fixation. Their short yet productive life cycles drive processes that drive carbon down to the ocean’s twilight zone and beyond, where it can be held for hundreds of years or more.
Recent studies of DNA viruses in the global ocean indicate that they play an intricate role in these essential ecosystem services. Viruses assume dual roles:
- as both assassin — killing up to 40% bacteria in the oceans every day
- and as collaborator — shuttling genes between hosts that can support key processes such as photosynthesis.
A statistical study of the RNA viruses collected during Tara’s voyage by experts at Ohio State University has revealed that RNA viruses infect and shuttle genes between eukaryotic plankton species .
Previous studies of DNA viruses have laid bare the role of viruses in influencing how ecosystems function, in evolutionary history, and even in the composition of the Earth’s atmosphere. For example, research exploring the impacts of bacteriophages in the global ocean have revealed that they kill up to 40 percent of bacteria across the ocean every day.
Paradoxically, this creates foundations for life.
By ripping bacteria to shreds, bacteriophages are thought to drive key nutrient cycles, releasing a molecular feast for marine organisms to eat. A proportion of these carbon-rich particles sink to the deep ocean, locking carbon away from the atmosphere, helping to buffer the effects of climate change.
Eukaryotic plankton also play a major role in processes such as photosynthesis and the carbon pump. In the new study, scientists have identified what they believe to be tell-tale genetic signatures of coevolution between RNA viruses and host organisms.
The research will support the identification of key ecological zones, help scientists understand the community structure and ecological drivers of RNA viruses, predict their hosts, and learn how these compare to knowledge of DNA viruses driving similar processes.
It lays the groundwork for investigating the scale of RNA virus diversity and the roles that RNA viruses play in influencing the life cycles of organisms crucial to nutrient cycles, oxygen production, and locking carbon away from the atmosphere.
To showcase background information : Piecing together viral puzzles
Massive questions about the biology of viruses in the ocean remained unanswered until scientists began tracking them in earnest just a few decades ago. Many of these puzzles scientists are still wrestling with today.
How many species are there? What underpins their diversity? What do they look like? What role do they play in their ecosystems? How have they influenced the evolution of life on our planet? Could we harness their secrets to our advantage?
To answer these questions, it is crucial to have access to data collected on a global scale in a systematic way. The Tara Oceans expedition returned a bounty of samples and environmental data from hundreds of different locations and depths from across the global ocean.
Using this resource, the Tara Oceans consortium sought to establish baseline inventories of plankton in the ocean – from viruses to fish larvae. Observations made using samples collected during Tara’s voyage have led to catalogues of microbial genes from both prokaryotes – such as bacteria and archaea – and eukaryotes – such as microalgae and zooplankton. Studies have also led to the discovery of more than 200,000 DNA viruses.
These catalogues led to follow-on studies to assess interactions and biology’s roles in critical ocean ecosystem services like sinking anthropogenic carbon.
Examples of research making use of ocean water samples collected during Tara’s expedition include:
The surprising ubiquity of giant viruses in the ocean
The pole-to-pole diversity of DNA viruses in the ocean
Plankton networks driving carbon export in the nutrient-poor parts of the ocean
The potential biogeochemical impacts of common ocean viruses
Illuminating virus-related proteins in the ocean
Patterns and ecological drivers of viral communities in the ocean
Finding the right ‘virocells’ to study
Ultimately, the work could expand knowledge about the role of viruses in ocean processes essential in oxygen production, carbon capture, major nutrient cycles, and much more.
It could also help researchers better understand viruses, providing the basic understanding necessary for research that could ultimately support responses to global climate change or our response to the next global pandemic.
Author : Adam Gristwood