[Tara Océans] Discovery of new viruses in the ocean: Mirusviruses
A new scientific discovery was described in the journal Nature dated 19.04.2023. The article reports on a recently identified group of DNA viruses abundant in the Ocean: mirusviruses. These complex organisms were characterized by a team led by the CEA, the CNRS, the Genoscope, the Pasteur Institute and other institutions, thanks to the important open data base established from samples collected during the Tara Oceans expedition (2009-2013).
Viruses infect plankton in the ocean
Scientists characterized a new group of DNA viruses which they named «mirusviruses». “Mirus“ is a Latin word which means strange, or even surprising. The name reflects the unusual evolutionary traits of these viruses which have very complex genomes, with hundreds of genes and many functions which are currently completely unknown.
This discovery is essential for the scientific community because we can now better understand the biodiversity of plankton on the surface of the Ocean and the importance of viruses in these ecosystems. It also sheds light on the evolutionary history of viruses, since mirusviruses are halfway between the herpes virus and the giant viruses, another distinct group of viruses present in the Ocean. This is a real opportunity to study the ecology and evolution of DNA viruses.
What are mirusviruses?
Mirusviruses are very abundant on the surface of the Ocean, from the equator to the poles, where they play a role in regulating microbial life by infecting unicellular cells with a nucleus (eukaryotes). They are very present in planktonic cells.
It is likely that mirusviruses play many roles in the complex balance of plankton. However, this first study focuses on the complex evolutionary history of these viruses, and many ecological and climatic points still need to be clarified.
Mechanism of viral infection of marine plankton
Being genetically close to the herpes virus, can mirusviruses be a danger to humans? The response from Tom Delmont, CNRS-Genoscope researcher, is simple: “If mirusviruses could infect humans, they would have been known for a long time! Even accidentally swallowing seawater is no risk for us”. These viruses cannot infect us.
Scientists do not yet know exactly which of the unicellular eukaryotes are infected by the virus, and their mechanisms of action are still unknown. “Much remains to be understood about these viruses. However, all of these new findings point to unicellular eucaryote infection, which rules out animals. Although the herpes virus and the mirusviruses share an evolutionary history, they are very distant and probably have very different functional strategies. In fact, our analyses show that functionally speaking, mirusviruses are much closer to giant viruses which also infect unicellular plankton cells.”
What is the difference between a DNA virus and an RNA virus?
The difference between DNA and RNA viruses is in the type of nucleic acids used by the virus for the storage of its genetic material: the genes of RNA viruses are made up of RNA and not DNA (like human cells for example).
Scientists know that RNA viruses and DNA viruses play an important role in marine ecology, particularly in the regulation of plankton. As far as DNA viruses are concerned, two major groups were already known. Mirusviruses form a third major group of DNA viruses that infect plankton on a large scale. The discovery of mirusviruses is the beginning of a new adventure since it gives the world scientific community the keys to detect and study the role of these viruses in many ecosystems, whether Ocean, lakes, rivers or on land.
DNA sequencing and phylogeny: a scientific approach that pays off
According to Tom, the discovery of these DNA viruses was a scientific adventure. “In 2019, our research team observed an unusual evolutionary signal in the tsunami of DNA sequencing data from Tara Oceans expeditions. It was a bit like criss-crossing a huge beach with a metal detector. But in our case, we were looking for new viruses. We started from a marker gene (an RNA polymerase – an enzyme that synthesizes RNA from the DNA template – present in most giant viruses) and completed a metagenomic exploration of plankton (sequencing technique and DNA analysis) based on its phylogenetic signal (study of kinship ties between different individuals). Our approach was therefore a cocktail of metagenomics and phylogeny. And we found an evolutionary treasure in the Ocean, something that didn’t match anything known! The bioinformatics work that followed enabled us to discover and then characterize the genomes of a major group of DNA viruses, which we named mirusviruses.
Tracing the evolutionary history of viruses: the phylogenetic study
What is a phylogenetic tree used for?
Phylogenetic studies help the scientific community to contextualize in the form of a tree the evolutionary history of living things. The leaves close to each other represent, in this study, viruses that share a recent common ancestor. And the very distant leaves represent viruses that only share a very distant common ancestor.
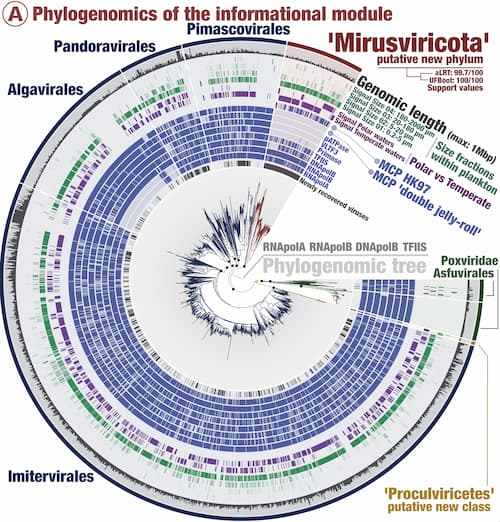
Mirusviruses form a new branch in this tree, distant from anything known to date. This is why they represent a major new group of DNA viruses.
“Viruses are not considered alive by a majority of scientists.” explains Tom. “However, we did discover an important component of the ocean biosphere. These viruses are probably present in every liter of water on the surface of the ocean! And we have characterized the genome of more than a hundred already. This group is likely to include thousands of virus species. Our latest results, which have not yet been published, certainly point in this direction. Mirusviruses are present in many ecosystems and they are very diverse.”
Identity card of mirusviruses halfway between giant viruses and herpes virus!
These viruses have an amazing history, halfway between the herpes virus, which infects half of the world’s human population, and the giant viruses, another group of viruses abundant in the Ocean and which has fascinated the scientific community for decades.
The genes responsible for the formation of the viral particle define the identity card of DNA viruses. And in mirusviruses, these genes have an evolutionary history close to that of the herpes virus. The origin of the herpes virus was an enigma, and these new findings help point the way to a planktonic origin hundreds of millions of years ago.
On the other hand, the majority of mirusvirus genes are similar to those of giant viruses. Mirusviruses are unique evolutive chimera that can help the scientific community clarify the evolutionary history of DNA viruses. “That is by far the most amazing aspect of this discovery.” confides Tom.
A discovery made possible by the Tara Oceans database
The Tara Ocean Foundation played a key role in this discovery,thanks to metagenomes generated at the Genoscope from samples collected during the Tara Oceans expedition.
For several years, Tom Delmont has specialized in the reconstruction and study of the environmental genomes of plankton, without going through cell culture. “I mainly rely on metagenomic data from Tara Oceans. Since 2019, I had the chance to coordinate an international team of researchers (including recognized experts in virus evolution) whose goal was to understand an unusual evolutionary signal found in Tara Oceans data. This signal was the starting point for discovering mirusviruses!”
Tara Oceans’ samplings have led to many other discoveries. Recently, our colleagues have characterized new groups of RNA viruses abundant on the surface of the Ocean. The discovery of these many RNA viruses is a fascinating story. An important trait RNA viruses and mirusviruses have in common — they infect unicellular plankton eukaryotes. Even so, they are very different: mirusviruses have very large genomes, whereas RNA viruses contain only a few genes and are much less complex. On the other hand, their evolutionary histories (between DNA viruses and RNA viruses) are very different, and therefore shed light on different aspects of the overall evolutionary history of viruses.
To follow-up this study, it will be necessary tocultivate these viruses in the laboratory. Several labs associated with Tara Oceans are already on this track. The majority of these viruses have not yet been characterized in the Ocean and beyond. Thus, Tom’s team will “strive to continue to explore the diversity of mirusviruses, and above all their fascinating genomic complexity, thanks in particular to the more recent expeditions led by the Tara Ocean Foundation.” To be continued!
Interview with Tom Delmont, CNRS-Genoscope researcher.
